Kinase-Mediated Cell Signaling Networks Control Cancer Cell Phenotypic Plasticity, Metastasis, and Therapy Resistance
The 518 protein kinases encoded in the human genome, i.e., the kinome, phosphorylate most proteins to alter their activity, localization, stability, and protein-protein interactions. Thus, kinases control most cellular processes and signaling pathways. Deadly human diseases, including cancers, neurodegenerative diseases, and immunological disorders, highjack the kinome to promote disease initiation and progression. Because of that, and because they are highly druggable with small molecule inhibitors, kinases have emerged as one of the most important classes of drug targets. Contrary to the classical, reductionistic views of cell biology, kinases engage in extensive crosstalk among one another and with other signaling enzymes, forming interconnected signaling networks. These networks are highly degenerate and robust towards perturbation like therapeutics, which drives therapy resistance in cancers and other diseases. Accordingly, network pharmacology approaches targeting multiple nodes in kinase networks are more likely to disrupt disease networks in an effective manner. We develop the next generation mass spectrometry (MS)-based proteomics technologies to systematically quantify how diseases alter kinome activity and connectivity, which is critical for prioritizing kinase networks for drug discovery (Project A). We have applied our novel technologies to identify kinase networks that control metastasis and therapy resistance in deadly hepatobiliary cancers like hepatocellular carcinoma (HCC) and intrahepatic cholangiocarcinoma (iCCA); these kinase networks may present future drug targets to combat deadly cancers (Projects B and C).
Targeting Oncogenic Endocytosis to Combat Liver Cancer Plasticity and Therapy Resistance
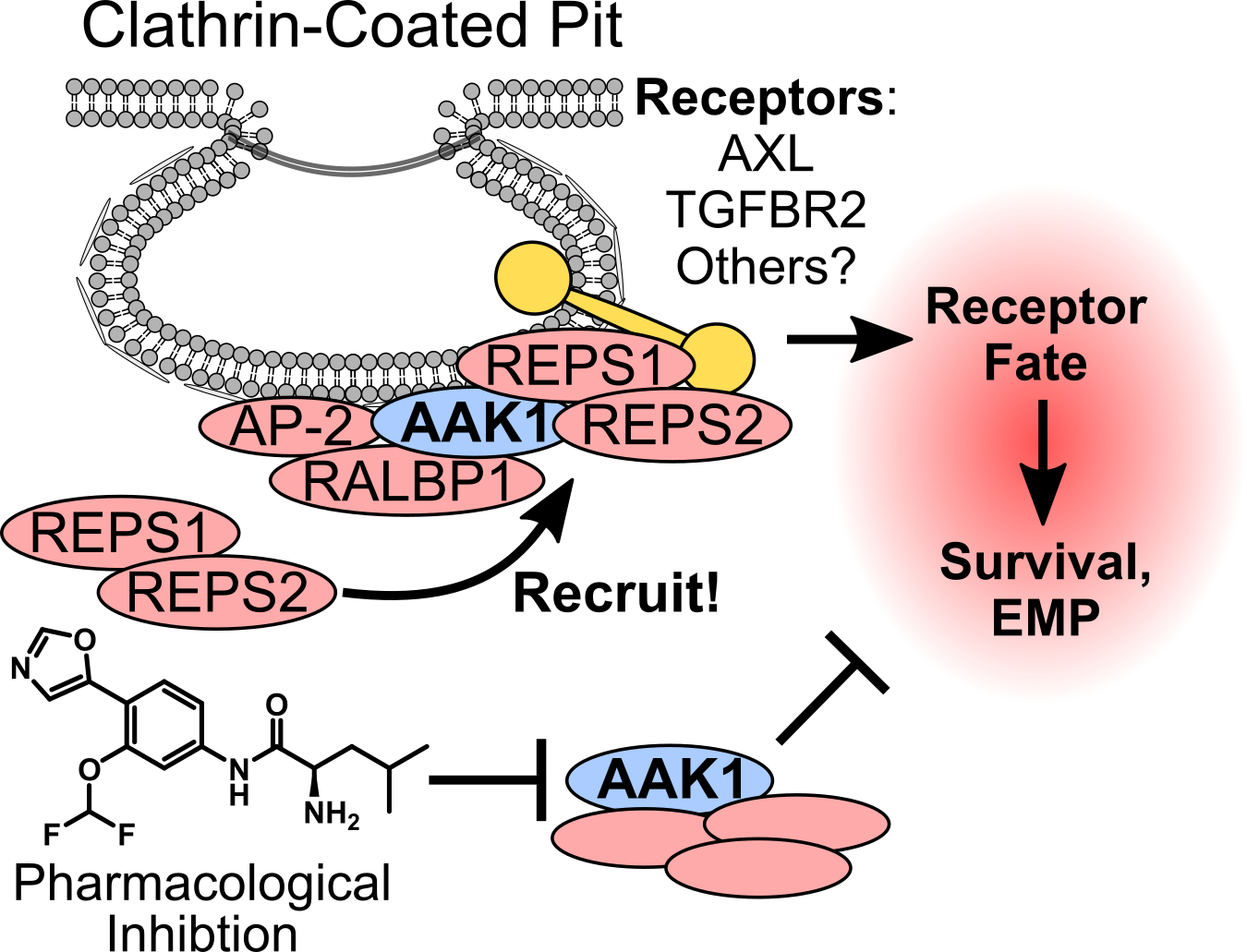
Hepatocytes, the cells of origin for most hepatocellular carcinomas (HCCs), are metabolic factories and membrane trafficking machines. We discovered that HCCs rewire the endocytic machinery to promote epithelial-mesenchymal plasticity (EMP), metastasis, and therapy resistance. Specifically, we identified an endocytic signaling complex controlled by the adapter-associated kinase 1 (AAK1) that is highly dysregulated in tumors. In this complex, AAK1 acts as a scaffolding protein that recruits specific endocytic adapters to alter receptor fates in favor of plasticity and survival. Accordingly, AAK1 and its’ interaction partners are novel drug target candidates to combat HCC metastasis and therapy resistance.
Targeting Plasticity and Cell Cycle Checkpoints to Induce Synthetic Lethality in Liver Cancers
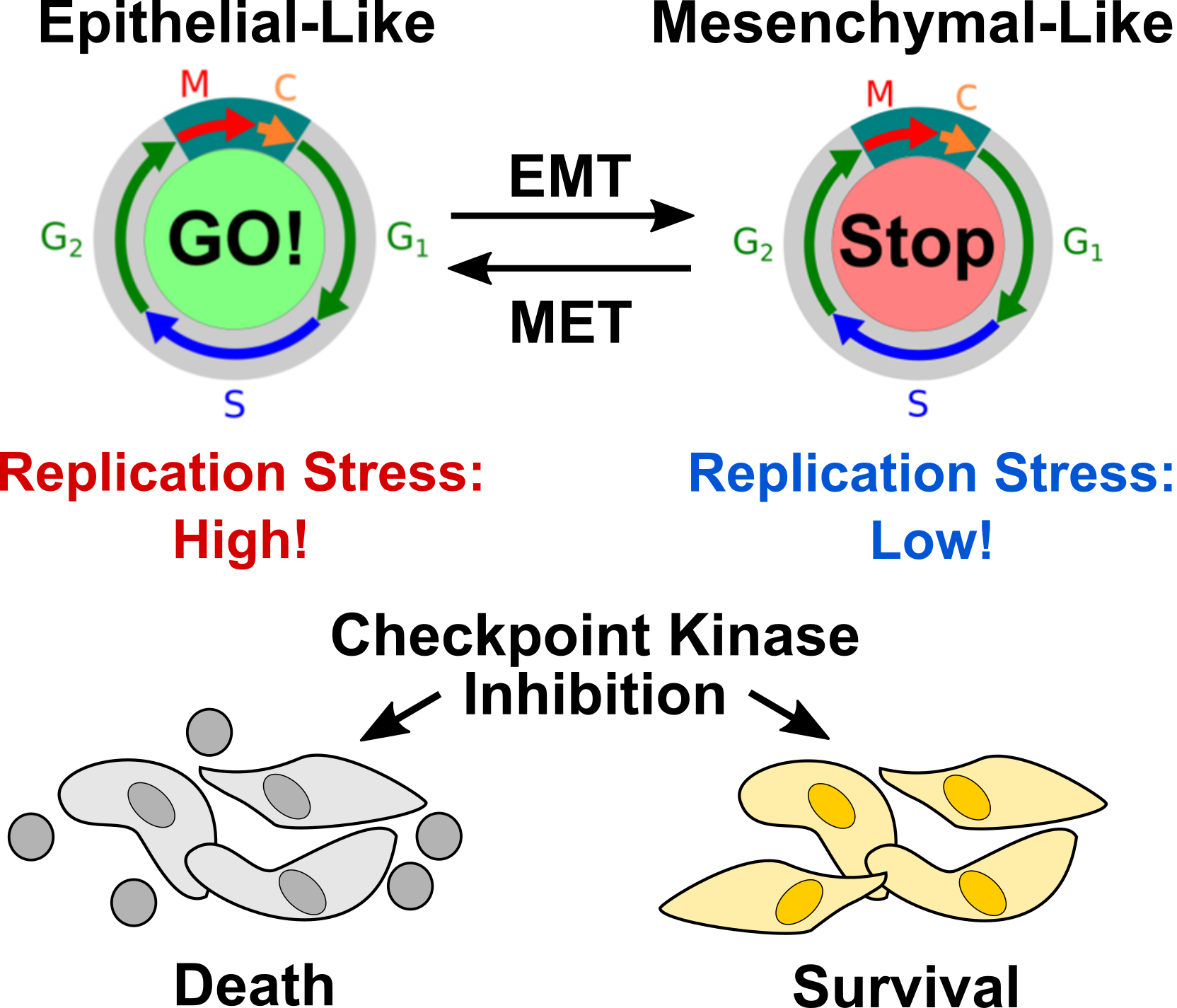
Most cancer cells suffer from replication stress (RS) due to the activation of oncogenic signaling pathways and the resulting hyperproliferation. Cancer cells activate DNA damage checkpoints to reduce replication stress and prevent premature entry into mitosis. In turn, inhibiting DNA damage checkpoints can drive cancer cells that suffer from RS into mitosis without resolving replication conflicts, causing mitotic catastrophe and cell death. We discovered that hepatocellular carcinoma (HCC) cells acquire epithelial-mesenchymal plasticity (EMP) to reduce RS, and that inhibiting EMP sensitizes HCC cells to DNA damage checkpoint inhibition, exposing an Achilles heel of HCC cells that can be exploited for novel combination therapies.
Developing MS-Based Proteomics Technologies for Multiplexed, Native Interactome Profiling
Protein-protein interactions (PPIs) are essential for all cellular functions and devastating diseases like cancers rewire PPI networks to drive disease progression and therapy resistance. Protein kinases are critical cell signaling enzymes that are deeply embedded in cellular PPI networks, altering the interactions of most proteins through dynamic and reversible phosphorylation. Accordingly, kinase complexes carry abundant information on disease states that can be exploited to identify disease mechanisms and therapeutic targets. We develop cutting-edge MS-based chemoproteomics technologies to systematically and rapidly map native kinase complexes.