27. Bryostatins 1 and 3 inhibit TRPM8 and modify TRPM8- and TRPV1-mediated lung epithelial cell responses to a proinflammatory stimulus via protein kinase C.
Sun L, Lamb JG, Niu C, Serna SN, Romero EG, Deering-Rice CE, Schmidt EW, Golkowski M, Reilly CA. Mol Pharmacol. 2025 Apr 22;107(6):100042. doi: 10.1016/j.molpha.2025.100042. Online ahead of print. PMID: 40378651
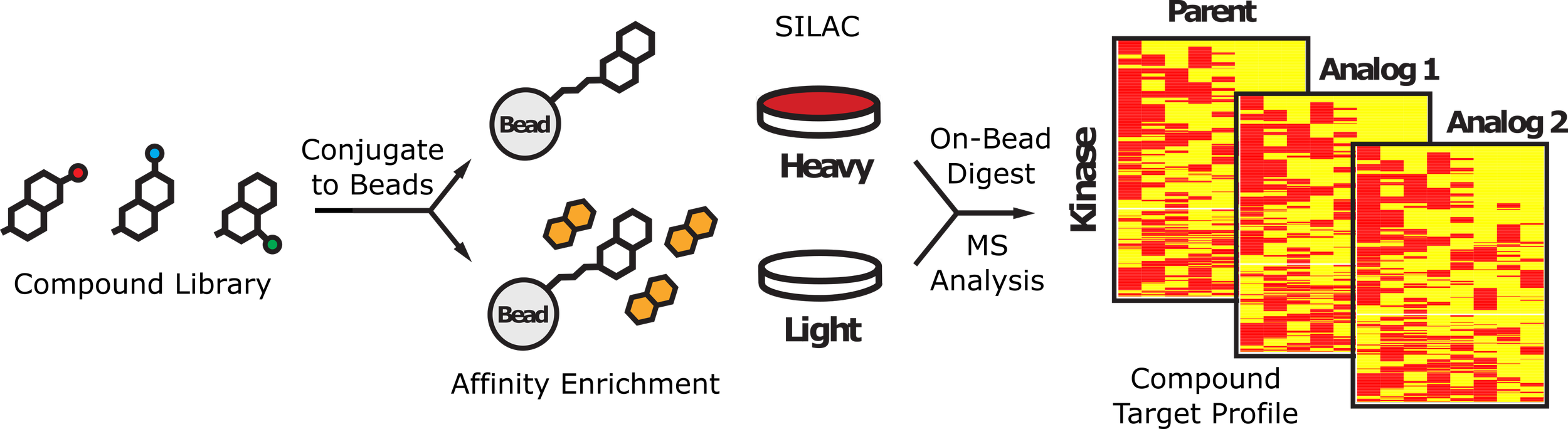
26. diaPASEF-Powered Chemoproteomics Enables Deep Kinome Interaction Profiling.
Woods K, Rants'o TA, Chan AM, Sapre T, Mastin GE, Maguire KM, Ong SE, Golkowski M. bioRxiv [Preprint]. 2024 Nov 22:2024.11.22.624841. doi: 10.1101/2024.11.22.624841. PMID: 39605566.
25. HAF Prevents Hepatocyte Apoptosis and Hepatocellular Carcinoma through Transcriptional Regulation of the NF-κB pathway
Acuña-Pilarte K, Reichert EC, Green YS, Halberg LM, Golkowski M, Maguire KM, Mimche PN, Kamdem SD, Hu PA, Wright J, Ducker GS, Voth WP, O'Connell RM, McFarland SA, Egal ESA, Chaix A, Summers SA, Reelitz JW, Maschek JA, Cox JE, Evason KJ, Koh MY. Hepatology. 2024 Sep 10. doi: 10.1097/HEP.0000000000001070. Online ahead of print. PMID: 39255518
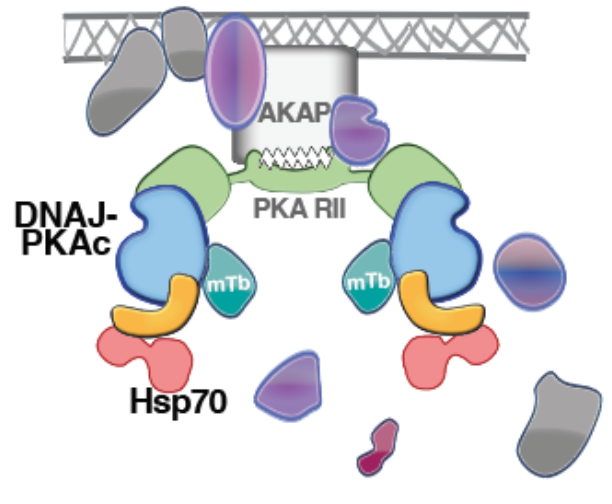
24. Recruitment of BAG2 to DNAJ-PKAc scaffolds promotes cell survival and resistance to drug-induced apoptosis in fibrolamellar carcinoma.
Lauer SM, Omar MH, Golkowski MG, Kenerson HL, Lee KS, Pascual BC, Lim HC, Forbush K, Smith FD, Gordan JD, Ong SE, Yeung RS, Scott JD. Cell Rep. 2024 Feb 27;43(2):113678. doi: 10.1016/j.celrep.2024.113678. Epub 2024 Jan 17. PMID: 38236773
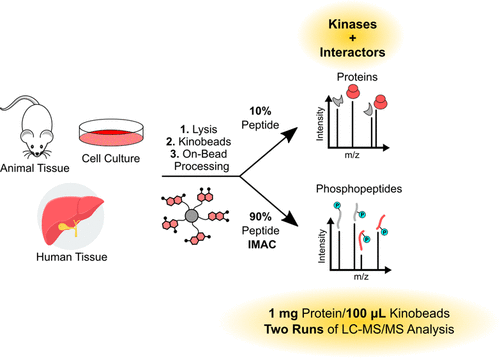
23. Multiplexed kinase interactome profiling quantifies cellular network activity and plasticity.
Golkowski M, Lius A, Sapre T, Lau HT, Moreno T, Maly DJ, Ong SE. Mol Cell. 2023 Mar 2;83(5):803-818.e8. doi: 10.1016/j.molcel.2023.01.015. Epub 2023 Feb 2. PMID: 36736316.
22. Proximity biotinylation to define the local environment of the protein kinase A catalytic subunit in adrenal cells.
Omar MH, Lauer SM, Lau HT, Golkowski M, Ong SE, Scott JD. STAR Protoc. 2023 Jan 4;4(1):101992. doi: 10.1016/j.xpro.2022.101992. Epub 2023 Jan 4. PMID: 36607814.
21. Mislocalization of protein kinase A drives pathology in Cushing's syndrome.
Omar MH, Byrne DP, Jones KN, Lakey TM, Collins KB, Lee KS, Daly LA, Forbush KA, Lau HT, Golkowski M, McKnight GS, Breault DT, Lefrançois-Martinez AM, Martinez A, Eyers CE, Baird GS, Ong SE, Smith FD, Eyers PA, Scott JD. Cell Rep. 2022 Jul 12;40(2):111073. doi: 10.1016/j.celrep.2022.111073. PMID: 35830806.
20. KiRNet: Kinase-centered network propagation of pharmacological screen results.
Bello T, Chan M, Golkowski M, Xue AG, Khasnavis N, Ceribelli M, Ong SE, Thomas CJ, Gujral TS. Cell Rep Methods. 2021 Jun 21;1(2):100007. doi: 10.1016/j.crmeth.2021.100007. Epub 2021 Jun 1. PMID: 34296206.
19. Phosphoproteomic Analysis as an Approach for Understanding Molecular Mechanisms of cAMP-Dependent Actions.
Beavo JA, Golkowski M, Shimizu-Albergine M, Beltejar MC, Bornfeldt KE, Ong SE. Mol Pharmacol. 2021 May;99(5):342-357. doi: 10.1124/molpharm.120.000197. Epub 2021 Feb 11. PMID: 3357408.
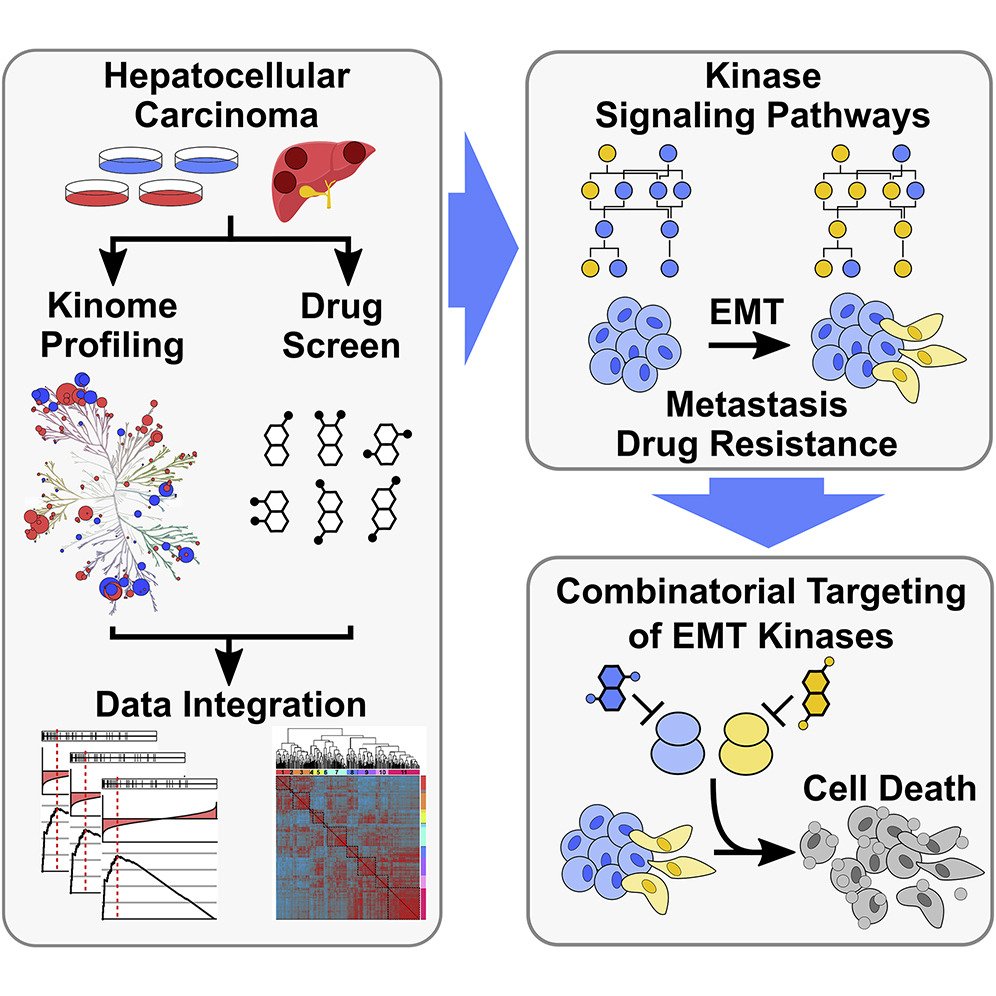
18. Pharmacoproteomics Identifies Kinase Pathways that Drive the Epithelial-Mesenchymal Transition and Drug Resistance in Hepatocellular Carcinoma.
Golkowski M, Lau HT, Chan M, Kenerson H, Vidadala VN, Shoemaker A, Maly DJ, Yeung RS, Gujral TS, Ong SE. Cell Syst. 2020 Aug 26;11(2):196-207.e7. doi: 10.1016/j.cels.2020.07.006. Epub 2020 Aug 4. PMID: 32755597.
17. Kinobead/LC-MS Phosphokinome Profiling Enables Rapid Analyses of Kinase-Dependent Cell Signaling Networks.
Golkowski M, Vidadala VN, Lau HT, Shoemaker A, Shimizu-Albergine M, Beavo J, Maly DJ, Ong SE. J Proteome Res. 2020 Mar 6;19(3):1235-1247. doi: 10.1021/acs.jproteome.9b00742. Epub 2020 Feb 27.PMID: 32037842
16. An acquired scaffolding function of the DNAJ-PKAc fusion contributes to oncogenic signaling in fibrolamellar carcinoma.
Turnham RE, Smith FD, Kenerson HL, Omar MH, Golkowski M, Garcia I, Bauer R, Lau HT, Sullivan KM, Langeberg LK, Ong SE, Riehle KJ, Yeung RS, Scott JD. Elife. 2019 May 7;8:e44187. doi: 10.7554/eLife.44187. PMID: 31063128.
15. Targeting Dynamic ATP-Binding Site Features Allows Discrimination between Highly Homologous Protein Kinases.
Chakraborty S, Inukai T, Fang L, Golkowski M, Maly DJ. ACS Chem Biol. 2019 Jun 21;14(6):1249-1259. doi: 10.1021/acschembio.9b00214. Epub 2019 May 13. PMID: 31038916.
14. A Combined Approach Reveals a Regulatory Mechanism Coupling Src's Kinase Activity, Localization, and Phosphotransferase-Independent Functions.
Ahler E, Register AC, Chakraborty S, Fang L, Dieter EM, Sitko KA, Vidadala RSR, Trevillian BM, Golkowski M, Gelman H, Stephany JJ, Rubin AF, Merritt EA, Fowler DM, Maly DJ. Mol Cell. 2019 Apr 18;74(2):393-408.e20. doi: 10.1016/j.molcel.2019.02.003. Epub 2019 Apr 4. PMID: 30956043.
13. Depletion of dAKAP1-protein kinase A signaling islands from the outer mitochondrial membrane alters breast cancer cell metabolism and motility.
Aggarwal S, Gabrovsek L, Langeberg LK, Golkowski M, Ong SE, Smith FD, Scott JD. J Biol Chem. 2019 Mar 1;294(9):3152-3168. doi: 10.1074/jbc.RA118.006741. Epub 2018 Dec 31. PMID: 30598507.
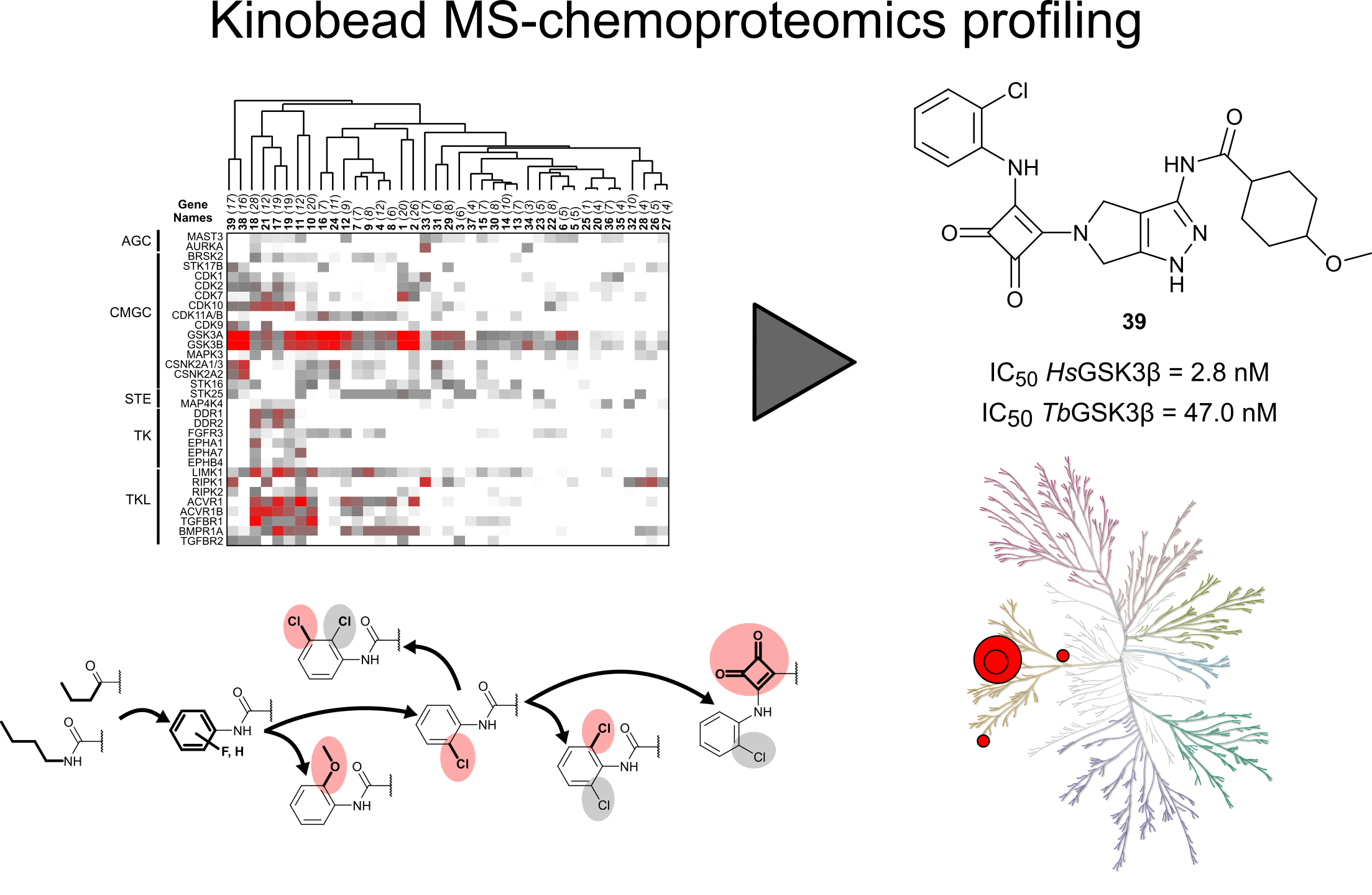
12. Kinome chemoproteomics characterization of pyrrolo[3,4-c]pyrazoles as potent and selective inhibitors of glycogen synthase kinase 3.
Golkowski M , Perera GK , Vidadala VN , Ojo KK , Van Voorhis WC , Maly DJ , Ong SE . Mol Omics. 2018 Feb 12;14(1):26-36. doi: 10.1039/c7mo00006e. PMID: 29725679.
11. 7 H-Pyrrolo[2,3- d]pyrimidin-4-amine-Based Inhibitors of Calcium-Dependent Protein Kinase 1 Have Distinct Inhibitory and Oral Pharmacokinetic Characteristics Compared with 1 H-Pyrazolo[3,4- d]pyrimidin-4-amine-Based Inhibitors.
Vidadala RSR, Golkowski M, Hulverson MA, Choi R, McCloskey MC, Whitman GR, Huang W, Arnold SLM, Barrett LK, Fan E, Merritt EA, Van Voorhis WC, Ojo KK, Maly DJ. ACS Infect Dis. 2018 Apr 13;4(4):516-522. doi: 10.1021/acsinfecdis.7b00224. Epub 2018 Mar 16. PMID: 29522315.
10. Proteomic Profiling of Protein Kinase Inhibitor Targets by Mass Spectrometry.
Golkowski M, Maly DJ, Ong SE. Methods Mol Biol. 2017;1636:105-117. doi: 10.1007/978-1-4939-7154-1_8. PMID: 28730476.
9. Analyses of PDE-regulated phosphoproteomes reveal unique and specific cAMP-signaling modules in T cells.
Beltejar MG, Lau HT, Golkowski MG, Ong SE, Beavo JA. Proc Natl Acad Sci U S A. 2017 Jul 25;114(30):E6240-E6249. doi: 10.1073/pnas.1703939114. Epub 2017 Jun 20. PMID: 28634298.
8. Kinobead and Single-Shot LC-MS Profiling Identifies Selective PKD Inhibitors.
Golkowski M, Vidadala RS, Lombard CK, Suh HW, Maly DJ, Ong SE. J Proteome Res. 2017 Mar 3;16(3):1216-1227. doi: 10.1021/acs.jproteome.6b00817. Epub 2017 Feb 3. PMID: 28102076.
7. SCAP/SREBP pathway is required for the full steroidogenic response to cyclic AMP.
Shimizu-Albergine M, Van Yserloo B, Golkowski MG, Ong SE, Beavo JA, Bornfeldt KE. Proc Natl Acad Sci U S A. 2016 Sep 20;113(38):E5685-93. doi: 10.1073/pnas.1611424113. Epub 2016 Sep 6. PMID: 27601673.
6. Spiro-fused carbohydrate oxazoline ligands: Synthesis and application as enantio-discrimination agents in asymmetric allylic alkylation.
Kraft J, Golkowski M, Ziegler T. Beilstein J Org Chem. 2016 Jan 29;12:166-71. doi: 10.3762/bjoc.12.18. eCollection 2016. PMID: 26877819.
5. Studying mechanisms of cAMP and cyclic nucleotide phosphodiesterase signaling in Leydig cell function with phosphoproteomics.
Golkowski M, Shimizu-Albergine M, Suh HW, Beavo JA, Ong SE. Cell Signal. 2016 Jul;28(7):764-78. doi: 10.1016/j.cellsig.2015.11.014. Epub 2015 Nov 28. PMID: 26643407.
4. Comparing SILAC- and stable isotope dimethyl-labeling approaches for quantitative proteomics.
Lau HT, Suh HW, Golkowski M, Ong SE. J Proteome Res. 2014 Sep 5;13(9):4164-74. doi: 10.1021/pr500630a. Epub 2014 Aug 12. PMID: 25077673.
3. Rapid profiling of protein kinase inhibitors by quantitative proteomics.
Golkowski M, Brigham JL, Perera GK, Romano GE, Maly DJ, Ong SE. Medchemcomm. 2014 Mar;5(3):363-369. doi: 10.1039/C3MD00315A. PMID: 24648882.
2. Strategy for catch and release of azide-tagged biomolecules utilizing a photolabile strained alkyne construct.
Golkowski M, Pergola C, Werz O, Ziegler T. Org Biomol Chem. 2012 Jun 21;10(23):4496-9. doi: 10.1039/c2ob25440a. Epub 2012 May 9. PMID: 22573264.
1. The 2-(triphenylsilyl)ethoxycarbonyl-("Tpseoc"-) group: a new silicon-based, fluoride cleavable oxycarbonyl protecting group highly orthogonal to the Boc-, Fmoc- and Cbz-groups.
Golkowski M, Ziegler T. Molecules. 2011 Jun 7;16(6):4695-718. doi: 10.3390/molecules16064695. PMID: 21654577.